L I L A B Bioinformatics and Genomics
Back to >> Software list VIcaller: Virome-wide clonal integration analysis VIcaller download Download VIcaller Version 1.1 and Users' Manual Note: we constantly update the software for new functions, fixed bugs, and others. If you would like to use the latest version, please send your email address to us (dawei.li at ttuhsc.edu) so that we can notice you when new versions become available. Questions? The software has been tested in multiple servers and by different users. If you have any questions about installation, error messages, or interpretation of results, feel free to contact the authors (dawei.li at ttuhsc.edu). Background related to the software In this method paper, we are introducing a novel strategy for discovering cancer viral etiology. Undiscovered viral etiologies in human cancers: Seven well-accepted oncogenic viruses can directly cause human cancers, collectively responsible for 12-15% of human cancers worldwide. Additional human viruses with oncogenic functions and virus-cancer associations remain undiscovered. Major challenges for identifying new oncoviruses: Virus-caused cancers usually have long latency after the original infection, from years to decades, and do not follow the Galilean principles of causality. It is difficult to prove oncogenic roles of identified viruses. However, the four most recently-identified oncoviruses (HCV, HPV, KSHV, and MCV) were found by searching viral nucleotide sequences in tumor tissues, demonstrating the historic successes of tumor sequence analysis. Whole-genome sequencing (WGS) provided novel opportunities for discovering unknown oncoviruses and new virus-caused cancer types. However, two major challenges have prevented previous studies from succeeding. First, viral sequence contamination is common in WGS data. Second, non-causal viruses may be present in tumors. The presence alone of a viral agent in tumor tissue cannot prove its oncogenic role nor distinguish it from non-causal viruses. Our strategy of searching for clonal viral integrations to overcome these challenges: The majority of virus-caused human tumors carry numerous viral integrations in their genomes. It provides a unique opportunity to overcome the two major challenges. First, identifying human-virus chimeric sequences flanking integration sites, rather than virus-only sequences, eliminates false discovery by viral sequence contaminations. Second, by further analyzing the percentage of cells with identical integrations, we can identify clonal integrations that occurred in the early stages of tumorigenesis. Similar to somatic mutations, viruses that lead to high cellular proportion “early stage” clonal integrations are potential oncogenic drivers. Therefore, analyzing clonal viral integrations in WGS data, as demonstrated in our research, is a very promising strategy for confidently determining new cancer viral etiologies (Fig. 1). 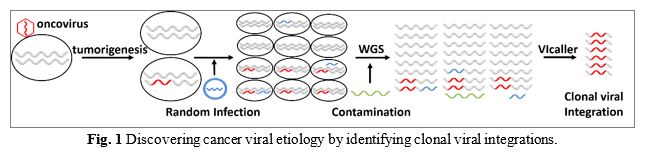 Our bioinformatics pipeline “VIcaller”: Our platform identifies oncoviruses by virome-wide screening for clonal viral integrations in cancer WGS data. By applying it into cancer genomic datasets, we have found genetic evidence for new virus-tumor association (such as high-risk HPVs in lung cancer) and previously undetermined oncoviruses (BK Virus in bladder cancer). The results demonstrate the capacity of our platform for identifying new cancer viral etiologies. Future applications: By applying our platform to the existing omics datasets, new cancer viral etiologies can be identified, including new oncoviruses and virus-tumor associations for cancer prevention. This study will provide strong genetic evidence for (1) additional human viruses that can cause cancers; (2) clear oncogenic roles of putative oncovirus candidates; and (3) additional tumor types that are associated with known oncoviruses. Oncovirus-induced tumors are mostly preventable. Demonstrating viral etiology will fundamentally change cancer epidemiology, as already seen in hepatocellular carcinoma and cervical cancer. Preventative approach, such as vaccination, may be developed for oncoviruses with significant health burden; and selection of at-risk population will allow for early detection of related-cancers. Oncoviruses also induce immune responses. With identification of more virus-associated tumors, potentially new application of the immune checkpoint blockade therapies may be developed. In all, we are introducing a novel strategy for uncovering new cancer viral etiologies, and we have demonstrated the capacity of our method in various cancer sequencing data. Citation We hope you find our tool useful! Please use the following reference: Chen X, Kost J, Sulovari A, Wong N, Liang WS, Cao J, Li D*. A virome-wide clonal integration analysis platform for discovering cancer viral etiology. Genome Research. 2019 May;29(5):819-830. PMID: 30872350. (* corresponding author) |
Copyright 2023 Texas Tech University Health Sciences Center The Li Lab.